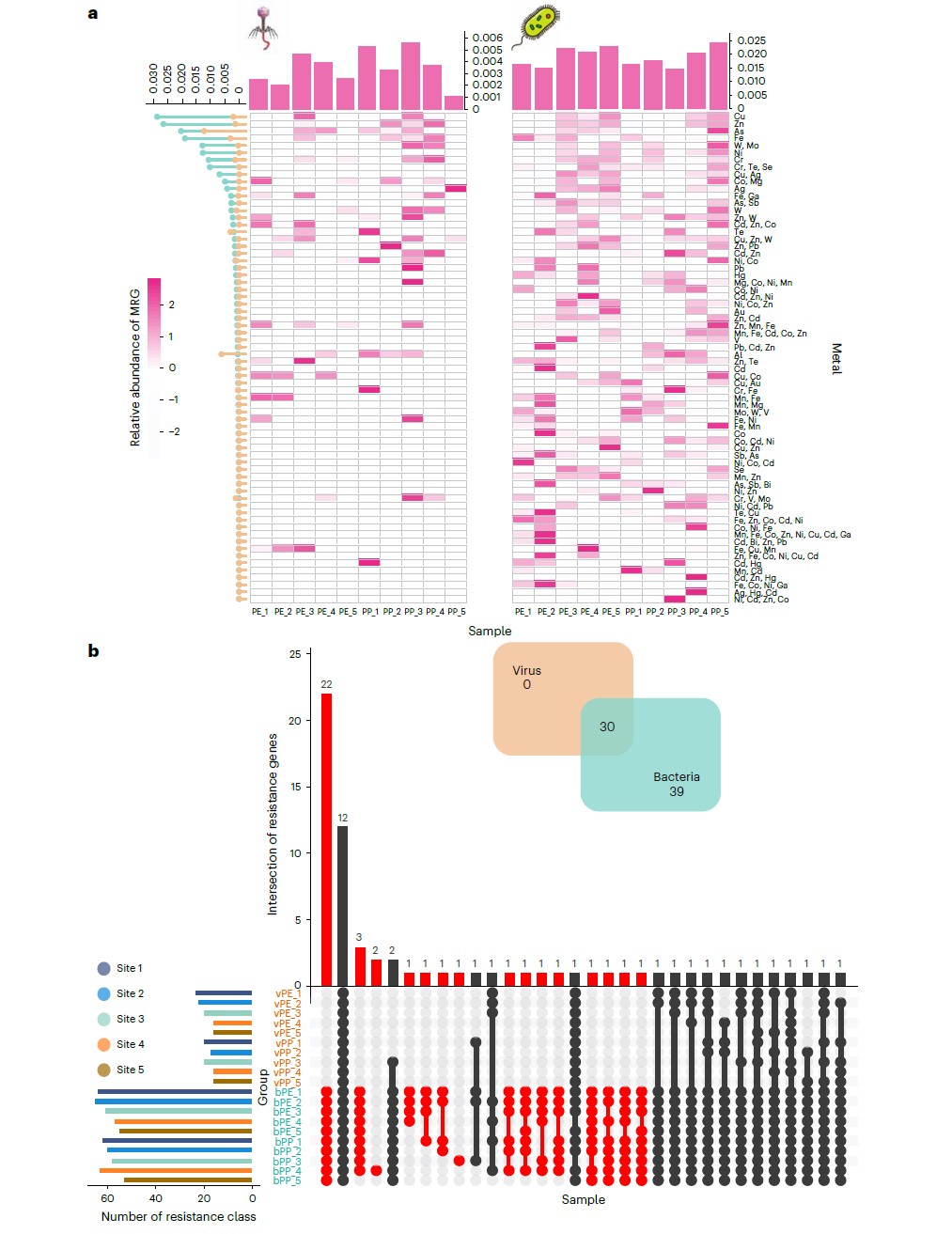
Microplastics provide a unique niche for viruses, promoting viral interactions with hosts and accelerating the rapid ‘horizontal’ spread of antibiotic resistance genes (ARGs). Currently, however, there is a lack of knowledge concerning the main drivers for viral distribution on microplastics and on the resulting patterns of viral biogeographic distributions and the spread of the associated ARGs. Here we performed metagenomic and virus enrichment-based viromic sequencings on both polyethylene and polypropylene microplastics along a river. Experimental results show that Proteobacteria, Firmicutes, Actinobacteria and Cyanobacteria were the potential hosts of viruses on microplastics, but only approximately 4.1% of viral variations were associated with a bacterial community. Notably, two shared ARGs and six metal resistance genes were identified in both viral and their host bacterial genomes, indicating the occurrence of horizontal gene transfer between viruses and bacteria. Furthermore, microplastics introduce more distinctive elements to viral ecology, fostering viral diversification and virus–host linkage while refraining from an escalated level of horizontal gene transfer of ARGs in contrast to natural matrixes. Our study provides comprehensive profiles of viral communities, virus-related ARGs and their driving factors on microplastics, highlighting how these anthropogenic niches provide unique interfaces that comprise highly defined viral ecological features in the environment.